Andrew White | EdisonScientific
Automating Scientific Discovery
Andrew White & Michaela Hinks
EdisonScientific
MIT NLP Seminar
December 2025
EdisonScientific
- Spinout from FutureHouse formed in 2025
- VC-backed AI Research Institute
- Based in San Francisco
- 35 employees
Science is changing independent of AI
Arxiv.org,10.6084/m9.figshare.17064419.v3
Number of Researchers are Growing
International R&D spending
PhD Researchers
NSF - https://ncses.nsf.gov/pubs/nsf24332; UNESCO UISI SDG9
Intellectual bottlenecks are growing
📝 Increasing paper count ($\approx$10M per year)
🧬 Larger data sets from cheaper
experiments (genome at
$200 per person, $1 / GB of sequencing)
🔍95% decline in disruptive papers since 1980
Park, M. et al. Nature 613, 138-144 (2023); Scannell, J.W. et al. Nat. Rev. Drug Discov. 11, 191–200 (2012); Deloitte 2025: Pharma innovation returns.
Edison Scientific Mission
Accelerate Scientific Discovery
Can large language models already do science?
LAB-Bench: Measuring Capabilities of Language Models for Biology Research
Jon M. Laurent, Joseph D. Janizek, Michael Ruzo, Michaela M. Hinks, Michael J. Hammerling, Siddharth Narayanan, Manvitha Ponnapati, Andrew D. White, Samuel G. Rodriques arXiv:2407.10362, 2024
Existing benchmarks
MMLU-Pro
- (Medicine) As of 2017, how many of the world's 1-year-old children today have been vaccinated against some disease?
- (History) On which continent are most of the Venus figurines found?
- (Chemistry) Find the logarithm of 3^2
Lab-Bench Questions
Not textbook knowledge
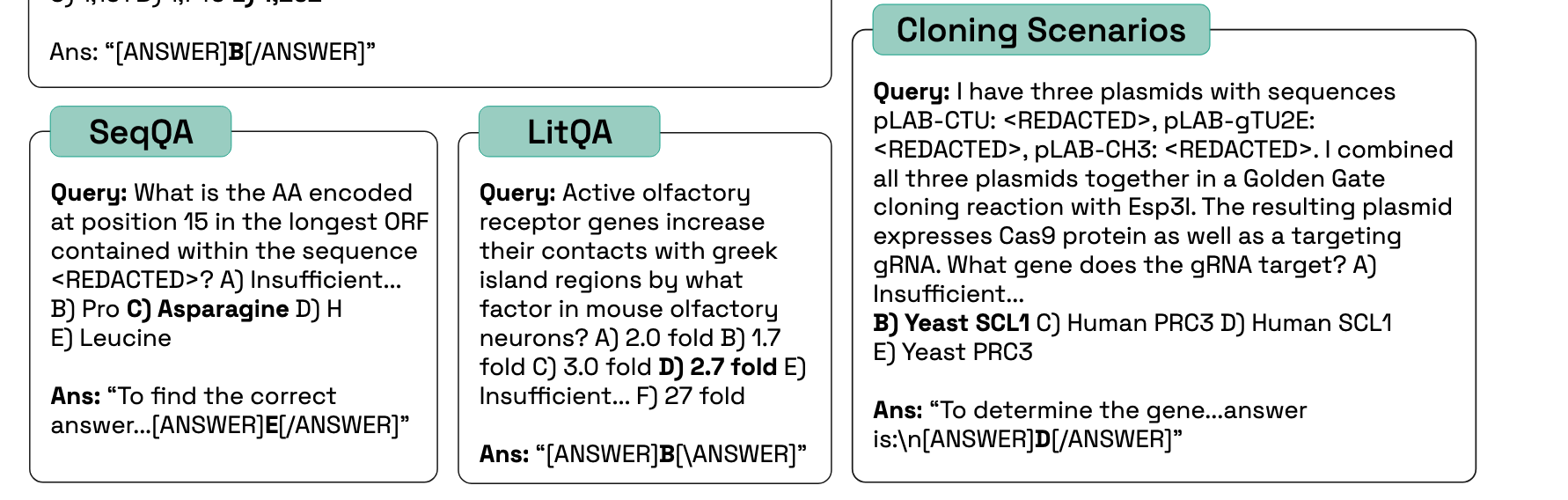
Lab-Bench Questions
Multimodal
Human baselines exceed LLMs
What about bioinformatics?
Bixbench Question:
How many peripheral immune cell types show significant differential expression (adjusted p-value < 0.05) of SOCS3?
Human baselines exceed LLMs
What is an agent?
Agent: trained, makes decisions
Environment: untrained, has tools, state
Protein Design Environment
- Protein design with 5 existing deep learning models
- Molecular dynamics, bioinformatics, literature research agent
- Input: "design 92 binders for PD-L1"
Wet lab validation
Learning vs Frontier Models
Crows
| Name | Environment | Key Tools |
|---|---|---|
| Literature | Literature Research | Search, Citation Traversal |
| Proteins | Designing novel proteins | AlphaFold2, Molecular Dynamics |
| Molecules | Designing new molecules | Retrosynthesis, self-driving robotic lab |
| Analysis | Generating discoveries from data | bioinformatics tools, code, file system |
Agent vs ML Model
Modify surface residues of IL-10 to increase expression and solubility in E. coli without disrupting dimerization or receptor interaction.
Automating research of scientific literature
Language agents achieve superhuman synthesis of scientific knowledge
Michael D. Skarlinski, Sam Cox, Jon M. Laurent, James D. Braza, Michaela Hinks, Michael J. Hammerling, Manvitha Ponnapati, Samuel G. Rodriques, Andrew D. White arXiv:2409.13740, 2024
Evaluating
Overexpression studies of PRMT4 in SW480 UPF1 knockout cells show that which arginine residue in PRMT4 is important for asymmetric di-methylation of UPF1 R433?
Better at answering questions than PhD biology experts
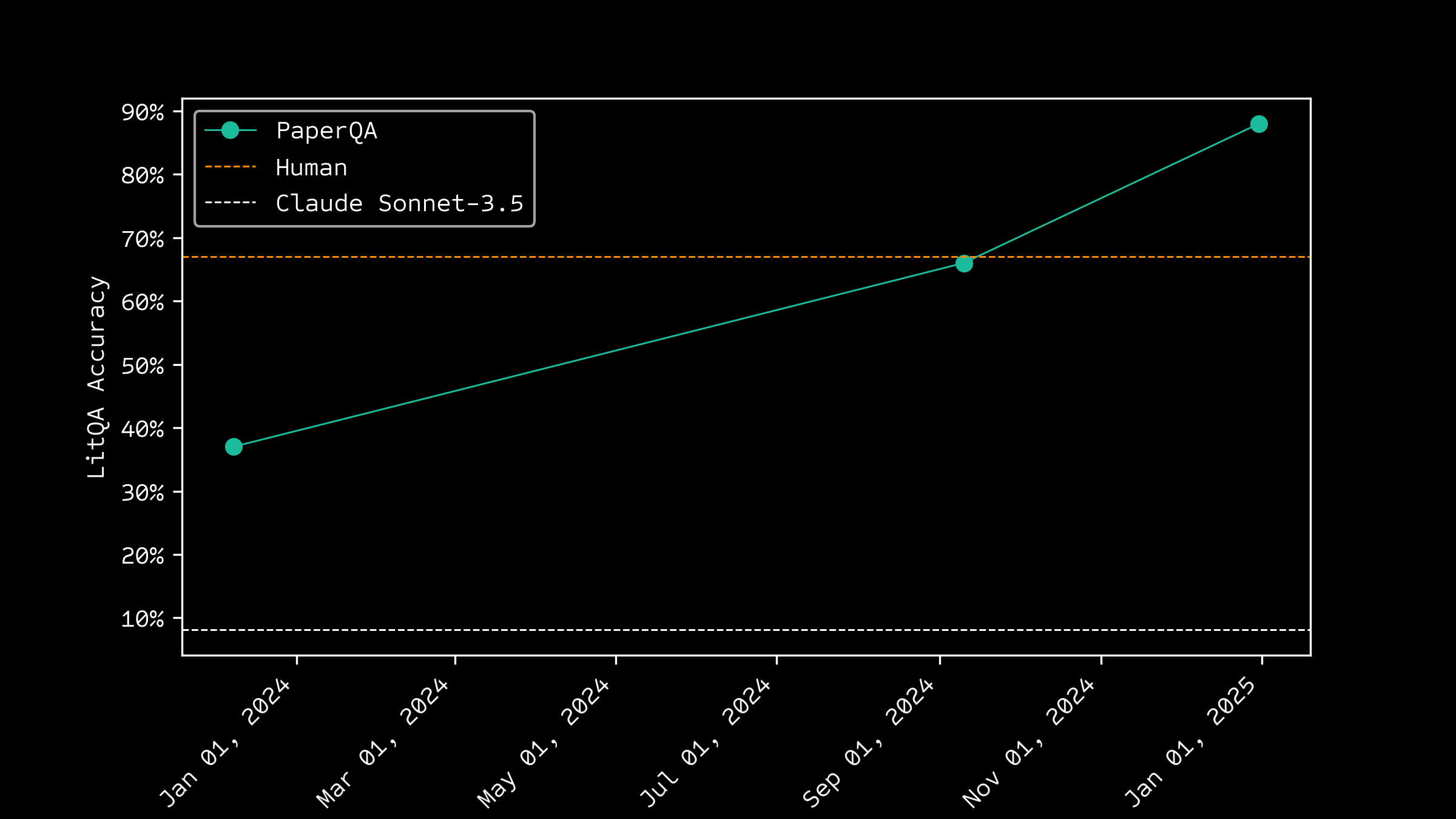
Improving over time
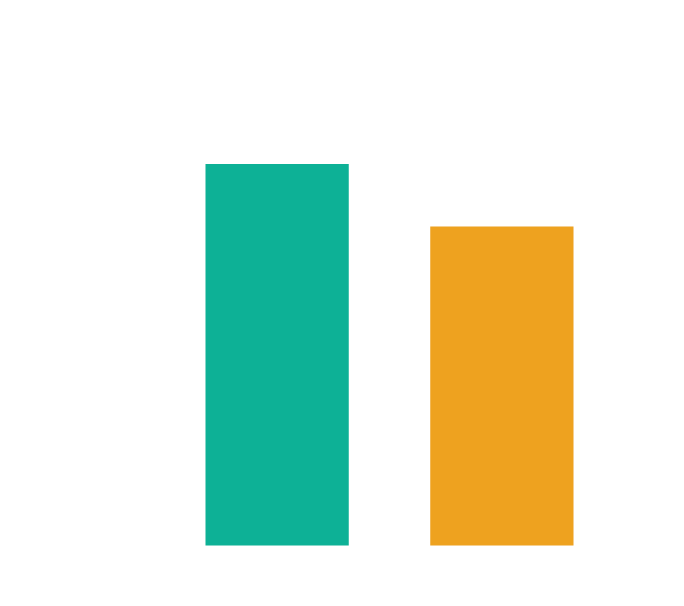
Better than human written Wikipedia articles
Can be used to check for precedent and disagreement in literature
Demo
Edison Platform
- Some amount of free usage, more for edu
- Grants for academic projects
- API - can be incorporated into your pipeline/agents
- Majority of agent code is open source
API
- Tasks per minute: 300
- Research Papers 175,000,000
- Wiki page for all diseases every 7 hours
- All arxiv papers per week 30,000 papers / month
- Check for contradictions 6.3M papers / year
- All Wikipedia every 3 weeks
Model intelligence will continue to increase
Complete cycle of disease to mechanism to target to drug
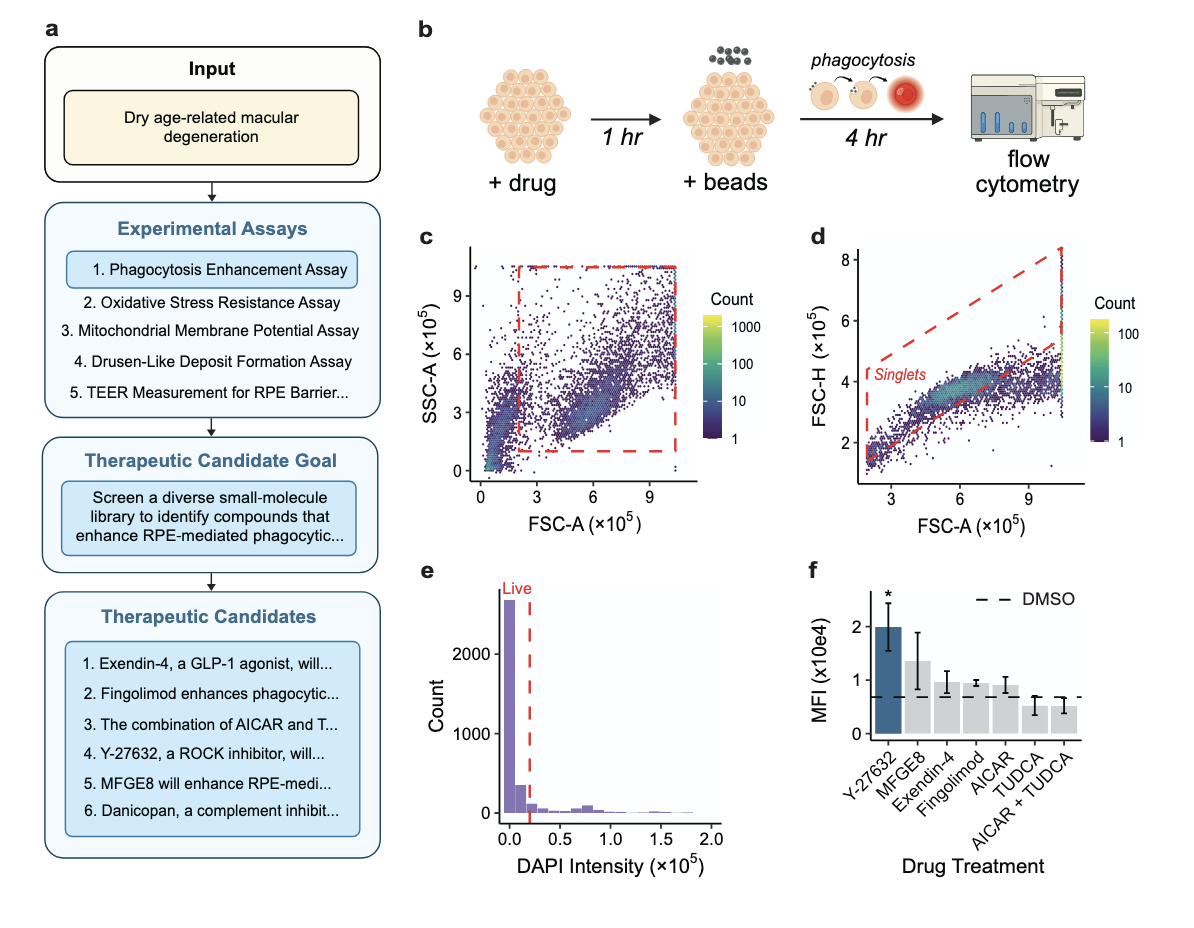
ROBIN: A Multi-Agent System for Automating Scientific Discovery
Ali Essam Ghareeb*, Benjamin Chang*, Ludovico Mitchener, Angela Yiu, Caralyn J. Szostkiewicz, Jon M. Laurent, Muhammed T. Razzak, Andrew D. White†, Michaela M. Hinks‡, Samuel G. Rodriques
Kosmos: An AI Scientist for Autonomous Discovery
Ludovico Mitchener, Angela Yiu, Benjamin Chang, Mathieu Bourdenx, Tyler Nadolski, Arvis Sulovari, Eric C Landsness, Daniel L Barabasi, Siddharth Narayanan, Nicky Evans, Shriya Reddy, Martha Foiani, Aizad Kamal, Leah P Shriver, Fang Cao, Asmamaw T Wassie, Jon M Laurent, Edwin Melville-Green, Mayk Caldas, Albert Bou, Kaleigh F Roberts, Sladjana Zagorac, Timothy C Orr, Miranda E Orr, Kevin J Zwezdaryk, Ali E Ghareeb, Laurie McCoy, Bruna Gomes, Euan A Ashley, Karen E Duff, Tonio Buonassisi, Tom Rainforth, Randall J Bateman, Michael Skarlinski, Samuel G Rodriques, Michaela M Hinks, Andrew D White
arXiv:2511.02824, 2025
Kosmos overview
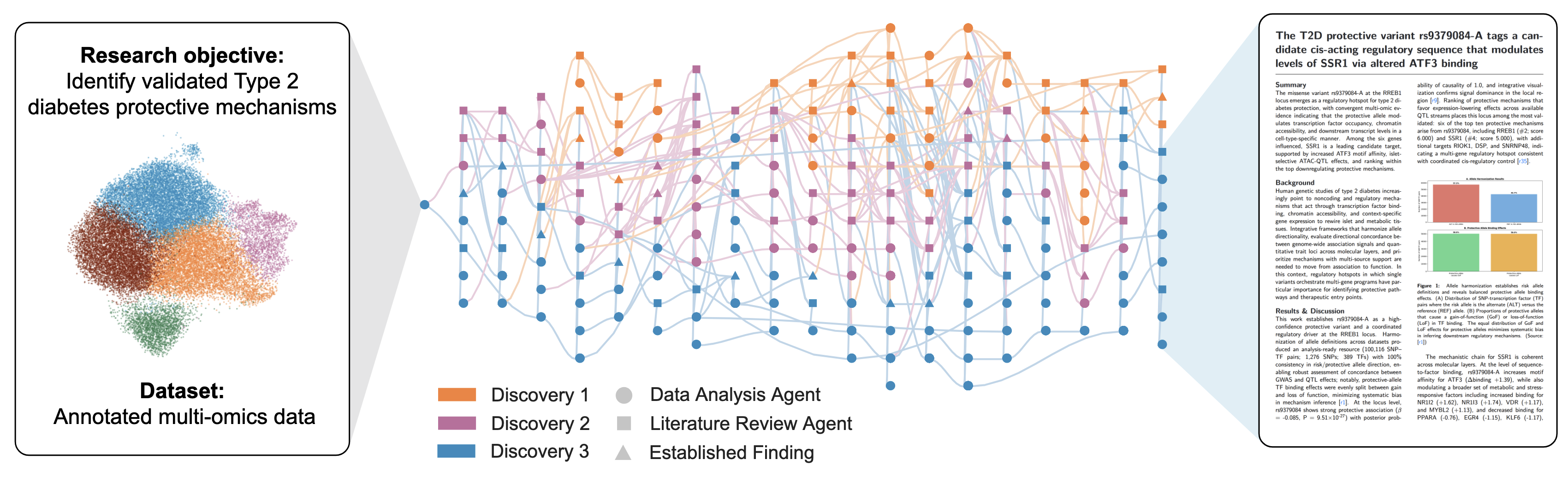
How do you validate systems like this?
Work with external groups. Input is their experimental data. Three discoveries reproduced in unpublished work. Four novel discoveries.

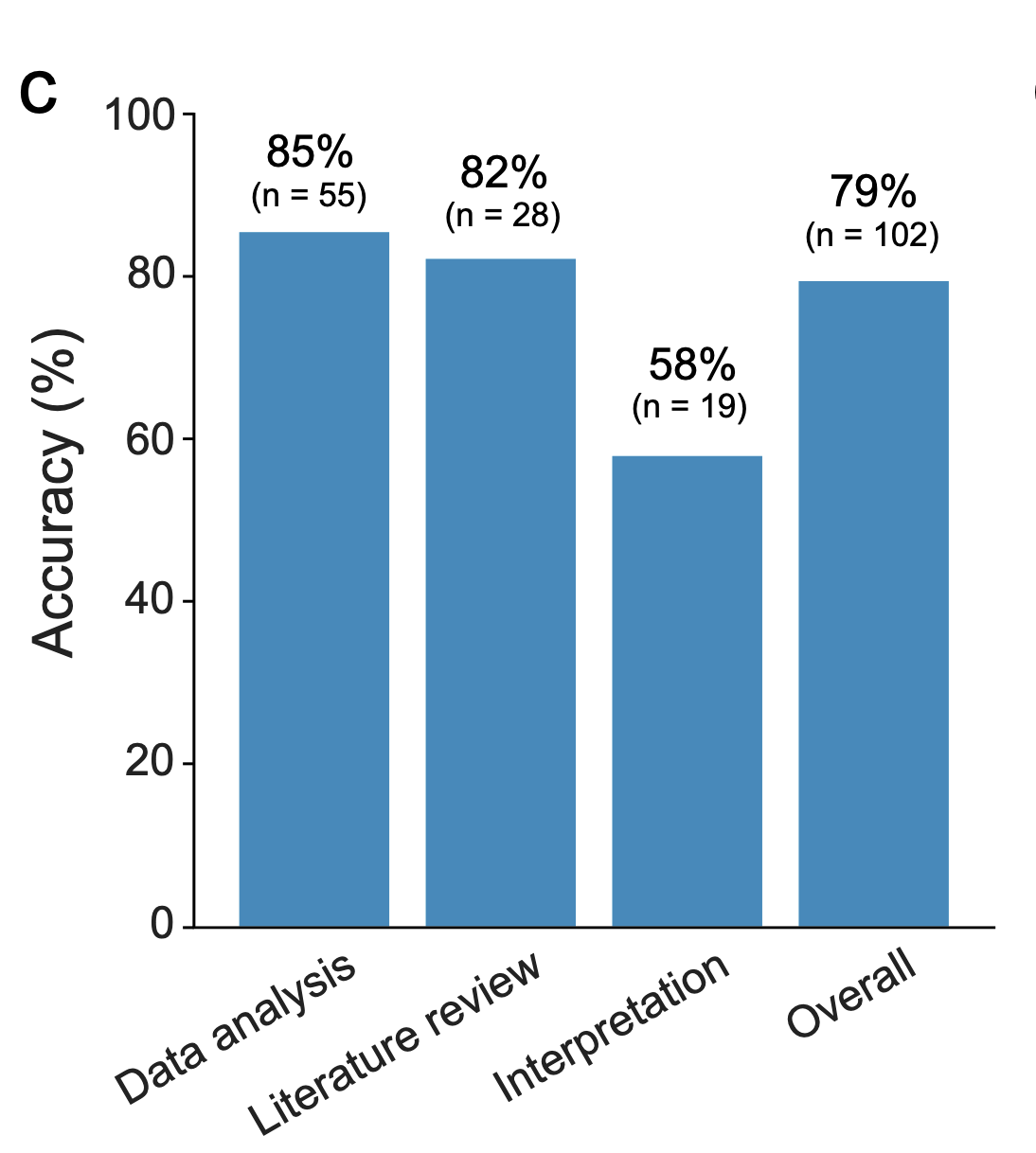
independent expert annotation of task difficulty and correctness
Example Discovery: What kosmos found
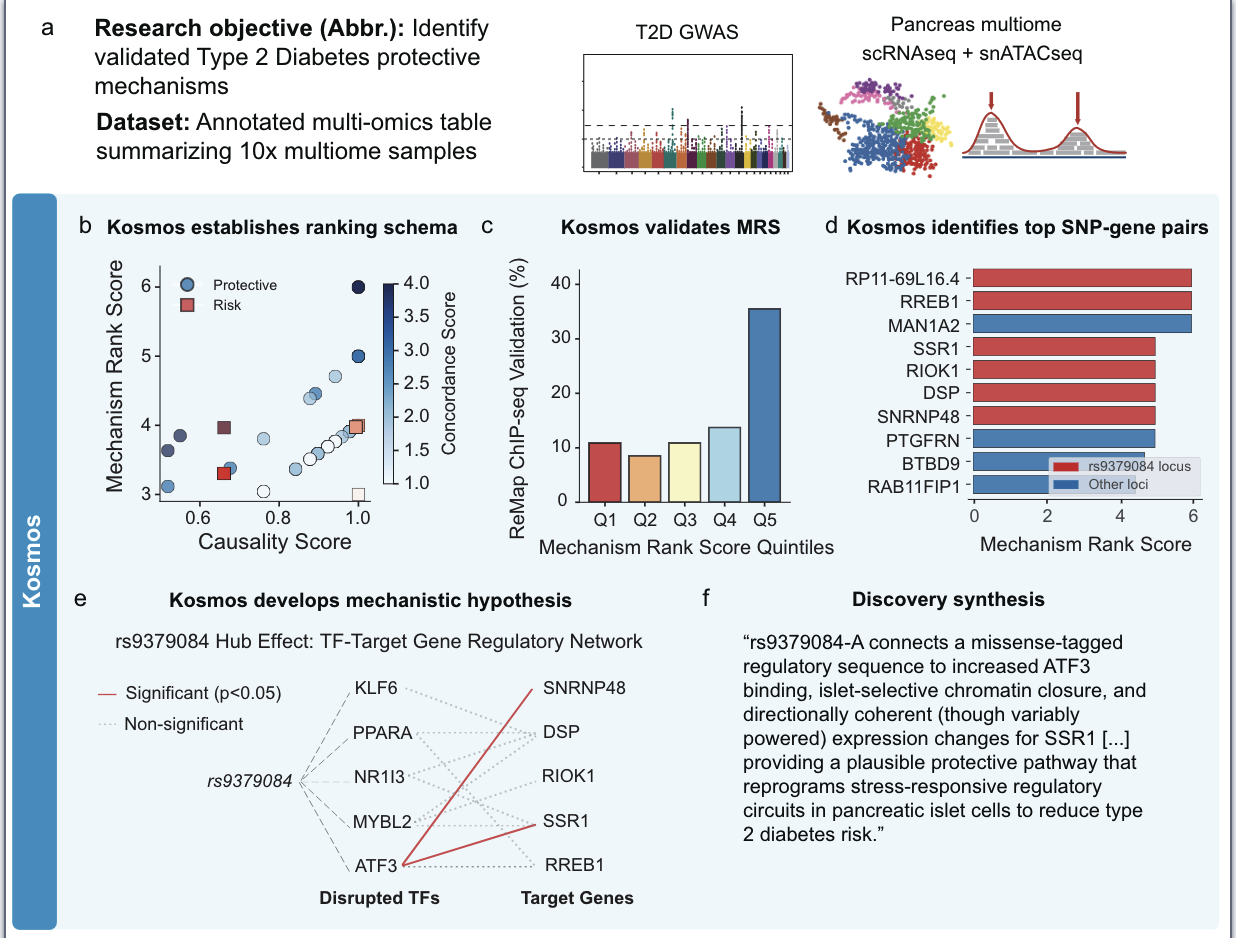
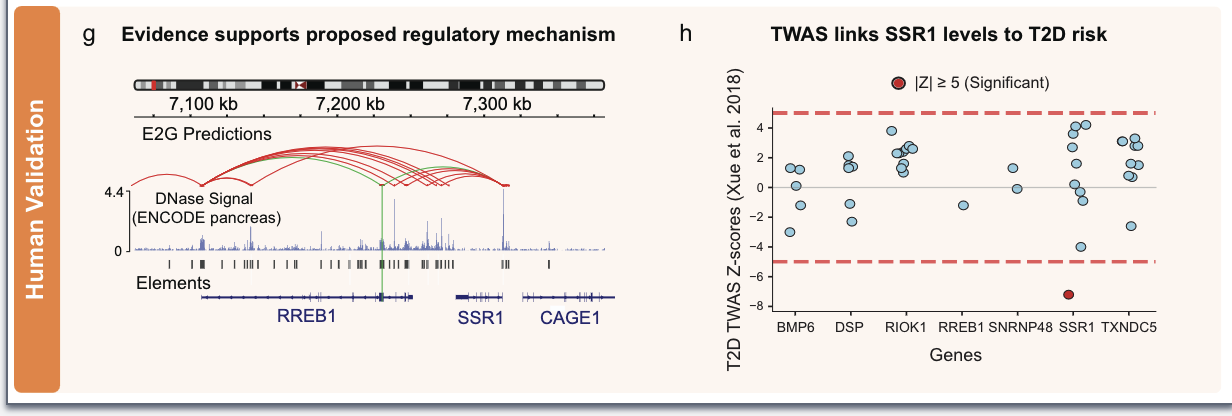
Human validation
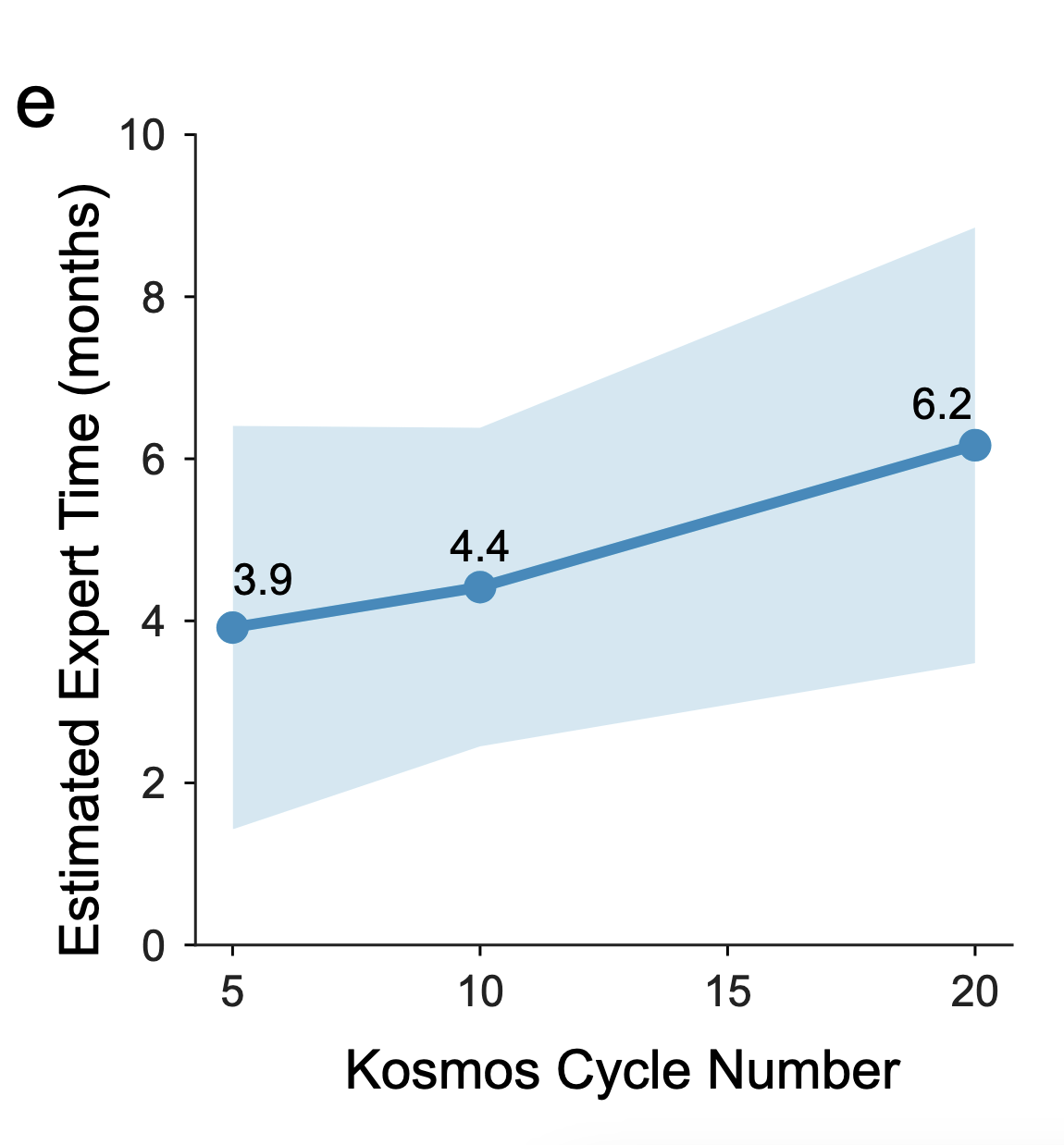
Kosmos Scale
- 120 sandboxed envs with 32GB RAM/8 CPUs
- 3,000 papers parsed and considered
- 24-48 hours of run time
- Generates up to 4TB of data
Try Kosmos Yourself
- 6 free runs with .edu email address
- Standard cost: $200 per run
- Available at platform.edisonscientific.com